Meta-Network
Meta-Network as an alternative framework to better represent the real species-species co-occurrence network. (Fig.1)Rooted in loose definitions of network as well as indirect correlation and mutual information, this framework outperforms other methods, as regard to network quality, in establishing and interpreting the species-species co-occurrence relationships, We have examined two large cohorts of microbial communities from human gut and ocean, and have established the species-species co-occurrence networks for respective samples. A number of key findings are presented in this work -- Firstly, the species-species co-occurrence network, generated by the loose definition strategy, contains more reasonable relationships among species. Secondly, the species-species co-occurrence patterns discovered by non-linear correlations and mutual information are functionally critical yet have not been identified by other existing approaches.
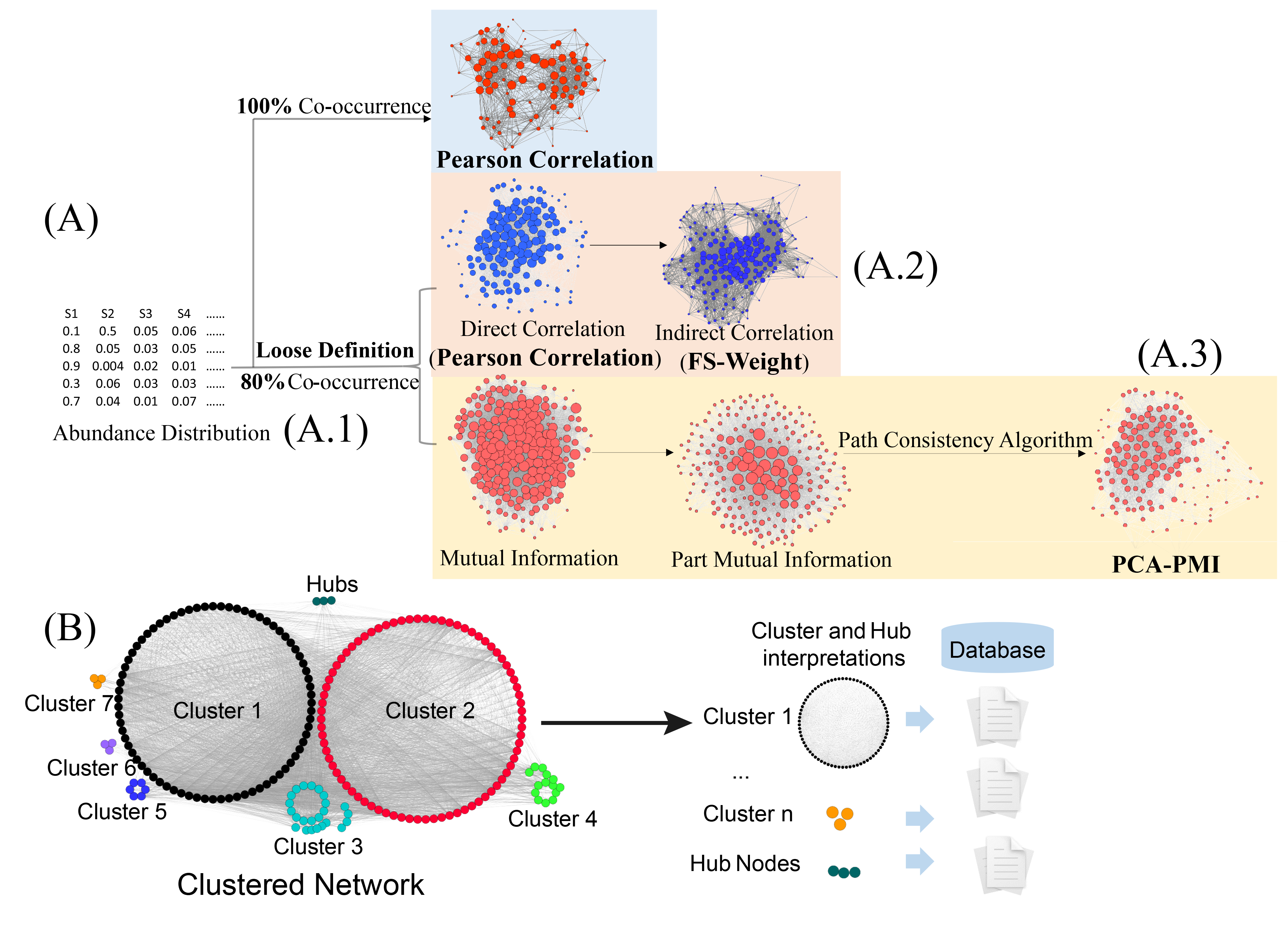
Fig.1. Network generation and cluster analysis for species-species co-occurrence network. (A) Network construction. Based on the loose definition method, networks were constructed by two methods (Fig 1, A.1). For detecting the indirect correlations, the direct correlations with Pearson or Spearman methods is calculated and a graph-based algorithm (FS-Weight) is applied to detect indirect correlations (Fig .1 A.2). For detecting the linear and non-linear correlations, the partial mutual information based on abundance distribution is calculated, then a few rounds of adjustment was perform based on Path Consistency Algorithm (PCA) until the network has no correlations changed (Fig 1, A.3). Node sizes represent the degree distribution. (B)Clustering analysis. Network clustering algorithm is applied to categorize the network into potentially functional or taxonomical enrichment units. Hub nodes represent species connected with many other species and usually play key roles in the community. Referenced databases and library investigation are used to identify the clusters and hub nodes.
Optimized Network Construction methods:
1.Loose definition of species-species relationship. We introduce the co-occurrence probability, a measures of the number of co-exist samples for pairwise species (Fig 1, A.1). To be precise, we first convert the original abundance matrix into a presence–absence matrix, and then calculate the co-occurrence probability (Fig 2.A). The correlation is calculated when two species have a co-occurrence probability over a user-defined threshold (Fig 2.B). It is worth noting that some genera pairs only exist in a few samples but have 100% co-occurrence in existing samples. These pairwise genera can yield abnormally high correlation from traditional Pearson and Spearman methods, but can be filtered out based on the sample quantity requirement with the loose definition method.

Fig 2. Loose definition method to find potential correlation. (A)Co-occurrence probability matrix for the sample data. Co-occurrence means two species have both non-zero abundance. The co-occurrence probability of two species is defined as co-occurrence times / total sample count*100%. (B)Network establishment with different co-occurrence thresholds. Pearson or Spearman correlations hold species with a 100% co-occurrence to calculate, but this threshold is too strict (left network). More correlations are detected with co-occurrence probability set to 80% (right network).
2.Direct and indirect relationships in the network. While direct relationships have been reported in many existing works in the context of intricate bacterial community, recent findings discovered plenty of indirect relationships which cannot be detected by the direct correlation 1. In our context, the FS-Weight method is applied to detect the indirect correlations (Figure1, A.2). FS-Weight measures the overlap between the interaction partners of two species, and is designed to estimate functional association between direct and indirect correlations.
3.Linearly and nonlinearly direct associations. We calculate PMI based on species abundance distribution. Since the order-one PMI cannot identify all the stable indirect correlations, we apply the Path Consistency Algorithm (PCA)3 to adjust the correlation distribution with a user-defined correlation threshold (0.2 used here). As a sparse problem with a large number of nodes 4, it is feasible to use PCA to construct the species-species network. We update the network after computing all the indirect neighbor correlations. Higher orders of PMI are calculated until there are no more edges changed.
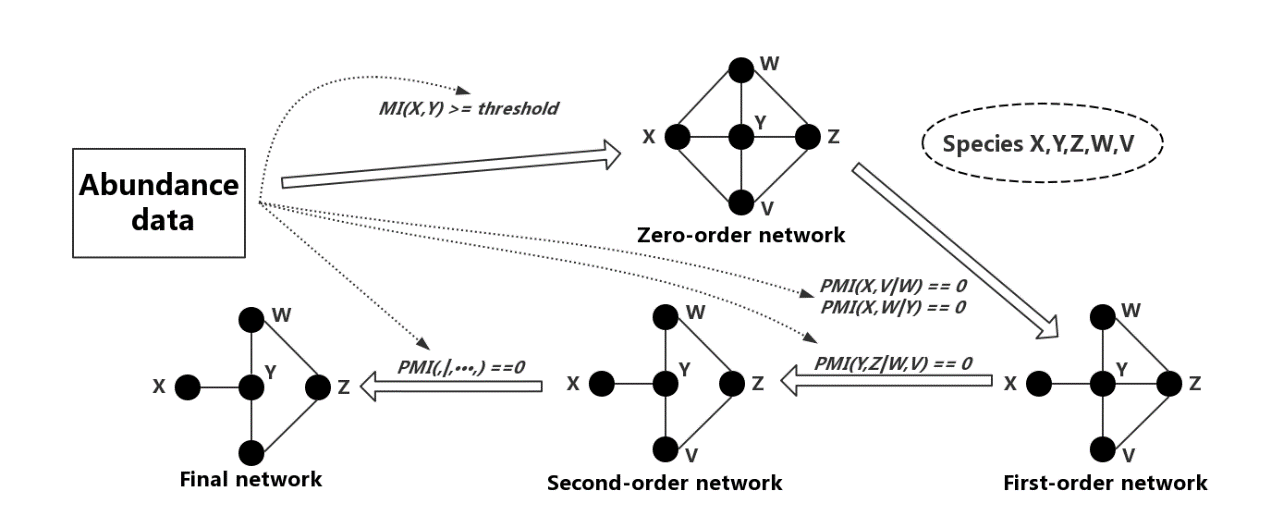
Fig.3, An overview of the PCA-PMI process. We first compute species abundance data by the mutual information method and then construct the zero-order network containing all the mutual information. We preform the PCA-PMI process to adjust the zero-order network. With a user-defined threshold, various orders of the PMI method are employed to assess whether the mutual information between two nodes was removed. When no more edges can be reduced, the network becomes stable.
4.Establishing clusters and hubs in the network We perform further analysis for cluster calculation and hub nodes detection based on Cytoscape (REF). We apply the MCODE cluster algorithm to detect the potential clusters, representing a set of species with similar or highly related functions (Wang, et al., 2015). Aligning cluster members against taxonomical or functional annotation databases can help us predict potential function or taxonomy of unknown species in the same cluster, cluster functions and taxonomical compositions. Further investigation to find important members in clusters are processed and identified as hub nodes, which connect with many with other nodes. After constructing a clustered species-species co-occurrence network, we measure the hub nodes based on degree distribution. The most connected nodes can be identified as candidates for hub nodes. These nodes are selected to perform further Kruskal–Wallis test to decide whether or not they are hub nodes.
Download :
Reference :
(Meta-Network referred to or integrated the following databases and tools.)
Price, M. N., et al. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol Syst Biol 9, 660 (2013).
Chua, H. N., Sung, W. K., Wong, L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 22, 1623-1630 (2006).
Zhou, C., Zhang, S. W., Liu, F. An ensemble method for reconstructing gene regulatory network with jackknife resampling and arithmetic mean fusion. Int J Data Min Bioinform 12, 328-342 (2015).
Zhang, X., et al. Inferring gene regulatory networks from gene expression data by path consistency algorithm based on conditional mutual information. Bioinformatics 28, 98-104 (2012).